.
INFINITY
The first and only English student magazine for medical laboratory sciences
No.5- August 2024
Director-in-Charge: Mahdieh Sadat Hosseini Nezhad
Editor-in-chief: Nazanin Zeinab Arefipour

Infinity:
No.5- August 2024
Concessionaire: scientific Association of Medical Laboratory sciences_varastegan Institute for Medical Sciences
ISSN: 320/8232
Director-in-Charge: Mahdieh Sadat Hosseini Nezhad
Editor-in-chief: Nazanin Zeinab Arefipour
Cover designer: Zeynab Mirnezhad
Editors: Mahdieh Sadat Hosseini Nezhad, Nazanin Zeinab Arefipour
Authors: Zeynab Mirnezhad, Hossein Rayat Pisheh, Melika Hosseinpour, Arezoo Ezanlo, Ali Ahmadi, Setareh Eskandari, Nastaran Tavabaki, Maryam Rohbakhsh Faalnezhad, Nazanin Zeinab Arefipour, Mahdieh Sadat Hosseini Nezhad
Editor-in-chief
When I see a new cell under the microscope, it feels like discovering a new planet through a telescope because, in the world of medical laboratory sciences, there's always a new planet waiting to be discovered and this infinite world plays a pivotal role in healthcare, encompassing disciplines such as clinical biochemistry, microbiology, hematology, immunology, genetics, and medical biotechnology. Each article featured in this magazine represents a critical exploration of disease mechanisms, diagnostic techniques, therapeutic strategies, and advancements in medical technology.
I invite you all to join us in this endeavor. Whether you're a student researcher eager to share your findings or a reader curious about the latest breakthroughs, Infinity is here to inspire and inform. Let's embark on this journey together, where curiosity meets discovery and each page of Infinity brings us closer to unraveling the mysteries of laboratory sciences.
Thank you to our dedicated authors and reviewers for their invaluable contributions.

Nazanin Zeinab Arefipour
Editor-in-Chief, Infinity
Director-in-Charge
Reaching the fifth edition of the only English-language journal in the field of laboratory sciences is a testament to the tireless efforts of the authors' team and all those who have supported us along the way.
In today's fast-paced world, we have always strived to contribute a small part to enhancing the knowledge and awareness of our country's scientific community in the laboratory sciences field by providing up-to-date and practical scientific articles.
Finally, we cordially invite all professors, students, researchers, and those interested in the life sciences to collaborate with us on future journal issues.
Sincerely,

Mahdieh Sadat Hosseini Nezhad
Director-in-Charge

Biochemistry
The role of Metformin in the fight against Colorectal Cancer

Hematology
-Antibacterial Capacity of chitosan in wound healing -Gene therapy for Hemophilia B with etranacogene dezaparvovec

Immunology
- Mesenchymal stem cells and their application in transplantation therapies

Microbiology
- A new antibiotic that has nothing to do with good bacteria

Genetics & Biotechnology
- Investigation of the Role of Effective Gene Therapy in Lung Cancer Control and Recovery
-Potential use of cancer cells as expression systems in pharmaceutical biotechnology
-Spatial Transcriptomics in Neuroscience and Neurological Disorders

News
.
part 1
Biochemistry
-The role of Metformin in the fight against Colorectal Cancer
The role of metformin in the fight against Colorectal Cancer
Introduction:
Colorectal cancer (CRC) remains a significant global health concern, with a high incidence and mortality rate. The disease arises from the uncontrolled growth of abnormal cells in the colon or rectum, often progressing slowly over several years. Early detection and effective treatment strategies are crucial for improving patient outcomes and reducing the burden of colorectal cancer on the healthcare systems.
Metformin, a widely prescribed medication for managing type 2 diabetes, has garnered attention for its potential role in cancer prevention and treatment. Studies have suggested that metformin may exert anti-cancer effects through various mechanisms, including the inhibition of cell proliferation, induction of apoptosis, and modulation of cellular metabolism. Its well-established safety profile, affordability, and widespread availability make metformin an appealing candidate for repurposing in oncology.
The fundamental mechanisms contributing to the protective effects of metformin in CRC:
Metformin has been shown to induce a temporary cell cycle arrest in colorectal cancer (CRC) cells. It effectively diminishes the expression of CRC stem cell markers, particularly CD44 and LGR5, thereby attenuating the stemness associated with CRC. This stemness is a critical determinant in the initiation, progression, and metastasis of colorectal cancer. Furthermore, metformin enhances the radiosensitivity of CRC cells and diminishes their capacity for DNA repair, even in P53-deficient CRC cells. It facilitates the differentiation of CRC cells while concurrently reducing their stemness. The drug induces cell cycle arrest in the G0/G1 or G2/M phases. Additionally, metformin amplifies the anti-cancer effects of various chemotherapeutic agents. It also inhibits DNA replication and proliferation in resistant CRC cells by downregulating the expression of the mini-chromosome maintenance protein complex (M-CM), which functions as a DNA helicase.
Research also indicates that metformin is capable of downregulating c-Myc and IGFR1 in colorectal cancer (CRC) cells. c-Myc plays a pivotal role in the reprogramming of gene expression in CRC, influencing various processes such as cellular proliferation, invasion, migration, immune evasion, chemoresistance, and the facilitation of the Warburg effect, which is defined by the process of aerobic glycolysis. The activation of c-Myc is mediated by the KRAS gene through downstream signaling pathways, including Wnt/β-catenin and MEK/ERK. Notably, cancer cells exhibit a preference for glucose fermentation to lactate over aerobic glycolysis, even when mitochondrial function remains intact. Furthermore, KRAS mutations are linked to the silencing of MATE1, a protein involved in the extrusion of multidrugs and toxins. The stimulation of IGFR1 enhances CRC cell proliferation via the PI3K/AKT/mTORC1 signaling pathway. Metformin exerts its effects by inhibiting critical proliferative pathways, namely PI3K/AKT, MEK/ERK, and Wnt/β-catenin, in CRC cells through an AMPK-dependent mechanism.
Epithelial-mesenchymal transition (EMT) plays a critical role in the migration and metastasis of cancer cells. Research indicates that metformin exerts an inhibitory effect on the metastasis of colorectal cancer (CRC) cells by interfering with the EMT process. Specifically, metformin reduces the expression of vimentin while enhancing the expression of E-cadherin, which collectively diminishes the invasive and metastatic capabilities of CRC cells. Additionally, vascular endothelial growth factor (VEGF) is recognized as a pivotal factor in intra-tumor angiogenesis, facilitating the growth and metastasis of CRC cells. Metformin has been shown to decrease VEGF expression in CRC cells. Furthermore, metformin also downregulates the expression of hypoxia-inducible factor 1α (HIF-1α) in these cells.
The other possible mechanism of action (MoA) of metformin is the inhibition of mitochondrial respiratory complex I, which leads to the activation of 5'-AMP-activated protein kinase (AMPK) in a serine/threonine-protein kinase STK11 (LKB1)-dependent manner. AMPK activation is associated with inhibition of the serine/threonine-protein kinase mTOR (mTOR), leading to favorable phenotypic outcomes in cancer cells, such as reduced protein synthesis and proliferation rates, activation of autophagy, and inhibition of inflammatory responses.
The advantage of using Metformin compared to other medications:
The research articles underscore the potential of metformin as a viable chemotherapeutic agent for the prevention of colorectal cancer (CRC), highlighting its safety profile, high patient adherence, cost-effectiveness, and well-defined mechanism of action. In contrast, non-steroidal anti-inflammatory drugs (NSAIDs), particularly COX-2 inhibitors, have demonstrated a significant reduction in CRC risk; however, they are associated with an elevated risk of major cardiovascular events. Metformin, a medication that has been utilized for several decades in the management of diabetes and other health conditions, was evaluated in a clinical trial. This trial revealed that low doses administered over one year resulted in minimal and mild adverse effects, thereby affirming its safety for prolonged use. The results indicate that metformin represents an economical and appropriate option for patients in need of long-term chemopreventive therapy.
Conclusion:
Collectively, both preclinical and clinical research indicates that metformin may play a significant role in the prevention and treatment of colorectal cancer (CRC), particularly in individuals at elevated risk, such as those with acromegaly or inflammatory bowel disease (IBD). Metformin has been shown to enhance the efficacy of various chemotherapeutic agents and to aid in overcoming chemoresistance associated with CRC. Furthermore, studies have demonstrated that metformin can substantially impede the development of new colorectal polyps and adenomas in non-diabetic individuals. Notably, even low doses of metformin (250 mg daily) have been associated with a decreased risk of colorectal polyps and adenomas.
References:
۱. Salovska B, Gao E, Müller-Dott S, Li W, Cordon CC, Wang S, Dugourd A, Rosenberger G, Saez-Rodriguez J, Liu Y. Phosphoproteomic analysis of metformin signaling in colorectal cancer cells elucidates the mechanism of action and potential therapeutic opportunities. Clin Transl Med. 2023 Feb;13(2):e1179. Doi: 10.1002/ctm2.1179. PMID: 36781298; PMCID: PMC9925373.
۲. Ala M. The Emerging Role of Metformin in the Prevention and Treatment of Colorectal Cancer: A Game Changer for the Management of Colorectal Cancer. Curr Diabetes Rev. 2022;18(8):e051121197762. doi: 10.2174/1573399818666211105125129. PMID: 34749618.

Zeynab Mirnezhad
Department of Medical Laboratory Sciences. Varastegan Institute for Medical Sciences, Mashhad, Iran

Mahdieh Sadat Hosseini Nezhad
Department of Medical Laboratory Sciences. Varastegan Institute for Medical Sciences, Mashhad, Iran
part 2
Hematology
-Gene therapy for Hemophilia B with etranacogene dezaparvovec
Antibacterial Capacity of chitosan in wound healing
The skin is a complex organ that plays a vital role in protecting the body from external environmental factors, regulating body temperature, and providing a physical barrier against water loss. It is the largest organ of the human body, representing approximately 15% of body weight [1]. The skin is composed of three main layers: the epidermis, dermis, and hypodermis, each with unique structures and functions. The outermost layer is the epidermis, which is primarily composed of keratinocytes and serves as the primary barrier against external environmental factors, such as UV radiation, chemicals, and microorganisms. The epidermis also plays a critical role in vitamin D synthesis [2-4]. The dermis is the second layer of the skin, which is composed of a fibroblast cell population. Fibroblasts are responsible for producing and secreting key extracellular matrix components, including collagen and elastin, which provide mechanical and elastic strength to the skin [6]. The dermis also contains other cell types, such as immune cells and sensory receptors, which play important roles in immune defense and sensory perception. The deepest layer of the skin is the hypodermis, which is composed of adipose tissue and connective tissue. The hypodermis plays a key role in thermal insulation by providing a layer of subcutaneous fat, which helps to regulate body temperature, and also acts as a mechanical barrier, protecting underlying tissues from damage [7,8].
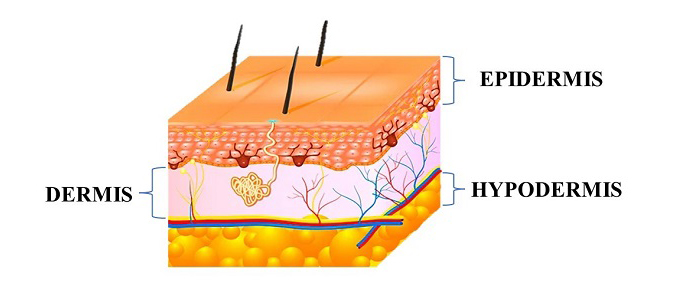
Wounds can be caused by physical injury, genetic disorders, or trauma [9,10,11]. Wound healing is a set of multifactorial and dynamic physiological processes. Different tissue repair processes consist of four stages coagulation and homeostasis, inflammation, proliferation, and regeneration. These four stages are time-dependent [12,13]. Local and systemic factors can influence the recovery process [14-17].
Wound healing results from interactions between cytokines, growth factors, blood, and extracellular network. Cytokines promote wound healing in various ways, such as stimulating the production of membrane components, preventing body dehydration, increasing inflammation, and creating granular tissue. This wound-healing process depends on various local and systemic factors. Some local factors include hypothermia (abnormally low body temperature), pain, infection, complete blood supply, and tissue oxygenation, which directly affect wound characteristics. And among systemic factors, we can mention stress, age, sex hormones, diseases such as diabetes, obesity, cancer, AIDS, smoking, and surgical technique [9,11]. Wound dressings are covers placed on the wound to protect the cells around the wound from environmental factors. In addition, wound dressings provide a suitable substrate for the growth and proliferation of cells. Wound dressings can partially fulfill the functions of damaged skin. Creating a barrier to prevent the penetration of bacteria and other factors that cause infection, keeping the wound environment moist, and allowing the penetration of gas molecules (permeability) are the most important features of a suitable wound dressing. The appropriate choice of a wound dressing depends on the characteristics of the wound, the information obtained from the wound, and the type of wound dressings available [12-15].
Biologically active natural materials have been used as potential materials in restoration due to their special properties. Due to their physical and chemical similarity, they can mimic the structure of human tissue. Natural polymers are more biocompatible than most synthetic polymers. Chitosan has been studied as one of the biopolymers, due to its unique properties, as a potential bioactive material. Chitosan is a derivative of chitin found in the exoskeleton of crustaceans, algae, and the bacterial cell wall [18,19].
Chitosan is distinguished from other biomaterials due to its unique properties: abundance, biodegradability, biocompatibility, hydrophilicity, non-toxic, antibacterial, antifungal, and wound healing effects [20]. It also has excellent elasticity, flexibility, and low inflammatory response [21]. The human body fully absorbs the products of the breakdown of chitosan (harmless amino sugars) [22-23]. These desirable properties accelerate the healing of various tissues and facilitate wound healing [24]. These unique properties have led to its use in some commercial products such as ChitoSamTM [25], ChitoGauze XR pro [26], ChitoFlex [27], and Axiostat® [۲۸]. However, the high molecular weight and high viscosity of chitosan and insolubility in most organic solvents and acid-free aqueous media limit it [29].
In this review, we will first evaluate chitosan's physicochemical and biological properties. Then we will examine the wound healing process and the use of chitosan in wound dressing based on research and patents made in recent years.
Chitosan
Rouget first boiled some chitin in potassium hydroxide in an experiment in 1859, which led to the discovery of chitosan [30]. Chitosan is a polymer in the family of linear polymers consisting of glucosamine and N-acetyl-D-glucosamine monomers. [31,32]. Also, the deacetylation reaction doesn’t cause the breakdown of chitosan chains [33]. Molecular weight and degree of deacetylation affect the product's pH, viscosity, turbidity, and temperature-sensitive properties [34]. The N-acetylated form of this polymer is called chitin. Chitin has a polysaccharide framework modified and amplified by a protein matrix [35]. Chitosan is a semi-acetyl (alkaline) form of chitin. In nature, the second biopolymer after cellulose is chitin [36].
Chitosan has a higher solubility than chitin. This difference in solubility causes the polysaccharide to be converted to a polyelectrolyte in an acidic environment [37]. Hydrophilicity and solubility of chitosan due to its free amino group, this material has a greater potential for chemical changes than chitin. Chitosan has three nucleophilic functional groups, including the C-2 NH2 group, the secondary C-3 OH group, and the primary C-6 OH group, present in each repeating chitosan molecule [38]. These functional groups in chitosan play a vital role in some physicochemical properties. Chitosan has derivatives consisting of hydrophilic and hydrophobic components. These derivatives result from modifications in the amino group or the hydroxyl groups. Chitosan has many special properties, including accelerating wound healing, antimicrobial, anticoagulant, antibacterial, antifungal, antitumor, anti-thrombogenic, film forming, hydrating agent, renewable, adsorbent, non-toxic and hemostatic [ 39-41]. It also can trap positive metal ions such as Ni2+, Zn2+, Co2+, Fe2+, Mg2+, and Cu2+. Of course, this capacity appears only in acidic conditions [42]. Today, chitosan has been considered by scientists and researchers and has been widely used in industries and biomedical products (fig.2). In the following, we will examine some unique properties of chitosan.

Sources & Extraction
In 1811, chitin or poly (β-(۱ → ۴) -N-acetyl-D-glucosamine) was discovered by Henri Braconnot [43]. Chitin is a type of biopolymer (polysaccharide) found in abundance in nature [44]. Chitin is present in many living organisms, such as insects (cuticles), crustaceans, and skeletal species [45-49]. Among the mentioned sources, shrimp and crabs are the most common sources as raw materials for preparing chitosan due to their low price and high chitin content. However, other sources, such as oysters, are also used [50- 53]. Different organisms contain different amounts (by weight) of chitin. For example, crustaceans have about 30 to 50 percent of calcium carbonate and 20 to 30 percent of weight of chitin. Still, Nephrop, which is a type of lobster in its shell, has about It weighs 60 to 75 percent by weight of chitin, which is the highest of all chitin-containing organisms [52, 54, 55]. Found in raw materials. Alkaline methods, including degreasing, degreasing, demineralization, and decolorization, are used to extract chitin [56].
The main derivative of chitin is chitosan (β- (l-4) -2-amino-2-deoxy-D-glucopyranose), which is a yellow linear polysaccharide containing the units N-acetyl-glucosamine and N-glucosamine [57]. The number of units of N-glucosamine is known as the degree of deacetylation (DDA) and plays an important role in chitosan. These units directly affect chitosan's physicochemical, biodegradable, and immunological properties [58]. The degree of chitosan deacetylation is measured by near-infrared spectroscopy, nuclear magnetic resonance spectroscopy, infrared spectroscopy, first derivative ultraviolet spectroscopy, etc [59].
Extraction of chitosan from chitin with different degrees of acetylation is done in two ways:
- Heterogeneous deacetylation
- Homogeneous deacetylation [60].
Both methods require concentrated alkaline solutions and a long processing time for acetylation. The processing time for the acetylation reaction may vary from 1 to 80 hours. Alternative processing methods, such as a set of thermal-mechanical processes using a low-alkaline cascade reactor [61], saturated steam flash operation [62], and the use of microwave dielectric heating [63]; And intermittent rinsing of water [64], have been created to reduce the processing time and a large amount of alkali. The most common processes available for the preparation of chitosan are mentioned.
As an alternative method, fermentation technology can also produce Chitosan from the fungal cell wall. Chitosan is found in the cell wall of a group of fungi such as Mucor rouxii [65], phycomyces and saccharomyces [66], and M. racemosus [67] [68,69]. The most important advantage of using these fungi to produce chitosan is that by controlling the fermentation parameters, its physicochemical properties can be manipulated [70]. Chitosan prepared from fungi showed less desirable and lower molecular weight, viscosity, and molecular weight than crustaceans. These desirable properties of chitosan prepared from fungi increase permeability [71,72].
Chitosan is obtained when the DDA ratio is more than 50%, and in most commercial chitosans, the DD range is between 70 and 90% [73]. Deacetylation is inversely related to molecular weight and directly associated with solubility in traditional solvents [74]. Due to the presence of the primary amino group, chitosan is not soluble in water but is soluble in dilute organic acids with a pH of less than 6.5. On the other hand, water uptake by chitosan is higher than cellulose. More water uptake in chitosan is due to the formation of polyelectrolytes from amino groups. One of the advantages of using chitosan instead of chitin is that chitin requires a toxic solvent such as dimethyl acetamide, while chitosan is readily soluble in dilute organic acids [75].
Antibacterial Activity
Bacterial infection is one of the most important and serious threats to human health [76]. Chitosan has a wide range of strong antibacterial activity, especially on gram-positive and gram-negative bacteria [77-81]. The level of antibacterial activity of chitosan and its derivatives are not the same; these compounds show different activity than gram-positive and gram-negative bacteria [82,83]. The cell wall in gram-positive bacteria consists of a thick layer of peptidoglycan. Teichoic acids are attached to N-acetylmuramic acid in this layer by covalent bonding. Teichoic acid affects the passage of ions through the outer surface layers inwards. Gram-negative bacteria, by the outer membrane (OM), cover a peptidoglycan layer above their cytoplasmic membrane. Important components of OM include lipoprotein and lipopolysaccharide (LPS). Due to the inactivity of hydrophobic compounds against gram-negative bacteria, the outer membrane barrier of gram-negative bacteria must be overcome to interact with the inside. One of the most important studies for using materials or scaffolding in wound healing is the study of its antibacterial properties. The antibacterial activity of a single material is evaluated by the disk diffusion method on two species of gram-negative bacteria (E. coli, P. aeruginosa) and one species of gram-positive bacteria (S. aureus). The microbial culture medium is prepared according to standard protocols as required. The microbial culture medium is then poured into 10 cm plates. A suspension of 0.5 McFarland bacteria is then prepared, and sterile cotton swabs are prepared. In the next step, these swabs are cultured evenly on the surface of the plate containing Müller-Hinton agar. Scaffolds are prepared by placing the samples in an incubator for 24 hours at 37 ° C. Finally, the barrier areas around the scaffolding are measured and calculated [142].
The antibacterial action of chitosan probably interacts with the surface of bacteria [84-86]. This interaction of chitosan with bacterial levels is generally expressed in four models:
- Adsorption of chitosan cationic groups with negatively charged components on the surface of bacteria by electrostatic bonding (electrostatic bonds vary according to the type of bacterial species. This mechanism is the most common interaction of chitosan and bacteria) [87,88].
- Formation of ionic bonds and changes in the permeability of the outer coating of gram-negative bacteria (Prevent the transfer of nutrients to bacteria) [89].
۳- Targeting intracellular microorganisms (interaction of chitosan with DNA through the holes of bacteria and fungi) [90].
۴- Chelation of metal ions on the surface of bacteria by chitosan amino groups [91].
The effectiveness of chitosan antibacterial properties is affected by various internal and external factors such as molecular weight, degree of polymerization, DDA, pH, temperature, pathogen, solubility, etc (fig.3) [92-95]. Also shown in many studies Position of positive charges plays a very important role in determining the amount of antibacterial activity [96-101]. Chitosan and its derivatives in the soluble phase have stronger and more effective antibacterial activity than the solid phase [102]. In one study, the antibacterial properties of chitosan derivatives (chitosan and maltose, fructose, glucose, and glucosamine) in solution were investigated, of which the chitosan-glucosamine derivative was more effective than other derivatives [103]. Also, the antibacterial activity of chitosan and its derivatives works optimally only when the pH is less than pKa [104].

Conclusion
Wound healing is a dynamic and complex physiological process to replace damaged or destroyed tissue layers and can lead to several irregularities. Many efforts and studies have focused on wound care and healing, emphasizing new treatment approaches and the development of wound management technologies. As a result, it is very important to choose a suitable material in the product’s structure that can accelerate the wound healing process and has no side effects or toxicity. Chitosan is a biopolymer with non-toxic, biodegradable, biocompatible, antibacterial, anti-inflammatory, and other unique properties. Chitosan and its derivatives are also soluble in water. Abundance, availability, easy extraction processes and the ability to be used in various support structures (such as hydrogels, bilayer membranes, and electrospinning) have also made this biopolymer an attractive and common material medical applications. According to the studies we reviewed, chitosan can promote wound healing and has become one of the most promising biopolymers to promote wound healing.
In addition to the unique properties of this biopolymer, the lack of favorable mechanical properties is the main drawback of using chitosan as a wound-healing material. Therefore, in the future, we will see studies to improve chitosan's performance and mechanical properties. Studies in this field can focus on the prospects for developing appropriate chitosan-based materials with optimal treatment protocols for clinical applications.

Hossein Rayat Pisheh
PhD of tissue engineering, Shiraz medical university, Shiraz, Iran
Gene therapy for Hemophilia B with etranacogene dezaparvovec
Introduction
Hemophilia B is a blood clotting disorder that causes easy bruising and bleeding due to an inherited mutation in the factor IX gene, resulting in a deficiency of factor IX. Its prevalence is lower than factor VIII deficiency (hemophilia A). Hemophilia B was first recognized as a distinct disease in 1952.
Many women carriers of this disease do not have any symptoms. However, an estimated 10-25 % of female carriers have mild symptoms. In infrequent instances, women may experience symptoms that are classified as moderate to severe. In the following, one of the ways to treat hemophilia will be discussed.
The US Food and Drug Administration (FDA) approval of the etranacogene dezaparvovec (Hemgenix) in November 2022 was a landmark event, both in hematology and gene therapy. This is the first FDA approval for a gene therapy for hemophilia, the first FDA approval for a liver-associated virus (AAV) vector-mediated gene therapy, and the first FDA approval for an AAV vector made in insect cells. Gene therapy for hemophilia has been in clinical development for over 2 decades The advancement of liver-targeted AAV gene therapy has transpired within a comparable temporal context. This approval sets the stage for multiple clinical development programs based on AAV-mediated gene delivery to the liver for indications ranging from plasma protein deficiencies to metabolic disorders to lysosomal storage disorders.
Nathwani et al reported the first clinical trial of scAAV-mediated gene transfer in hemophilia B in 2011. This trial showed that the carrier was well tolerated and resulted in a dose-dependent increase in FIX activity levels. All patients showed stable FIX expression long-term, with mean FIX levels ~5% of normal in the high-dose group. These participants observed stable and therapeutic FIX expression over 10 years (as reported in 2022) without late toxicity.
Subsequent development of a rabbit model recapitulated the transporter findings in seminal fluid and demonstrated that transporter clearance is time- and dose-dependent. This risk is now typically reduced by using barrier contraception, ideally up to 3 consecutive sperms. The samples show measurements below detectable levels of carrier DNA in semen.
Risk of insertional mutagenesis
Several clinical studies have shown that AAV vectors can integrate into the target cell genome at a low frequency and that rAAV integrity depends on pre-existing double-strand breaks in the genome. The frequency of AAV integration is low, and given that no cases of rAAV-associated cancer have been reported in humans, the risk of malignancy appears to be theoretical.
Analysis of human biopsies after AAV gene transfer has confirmed that AAV integrity is low in abundance. If HCC development is driven by AAV vector integration, frequent integration events, with at least 1 dominant event, can be expected. History of hepatitis virus infection and old age are risk factors for HCC. In conclusion, combined molecular and vector analysis of the HCC index case after liver-based rAAV-based gene therapy determined that AAV is unlikely to be involved in HCC.
Conclusion
As a result, the lack of observed tumorigenesis associated with rAAV in large animals and humans suggests that this risk is low compared to neonatal mice. If the mouse data are indeed extrapolated to humans, the most provocative clinical trial for the potential mutagenic risk in humans is shown by Zol-gensma (onasemnogene abeparvovec-xioi), an AAV therapy approved for the treatment of spinal muscular atrophy. Zolgensma is given at a very high dose (the recommended dose is 1.1 x 1014 carrier genomes per kilogram of body weight administered as an IV infusion) to pediatric patients under 2 years of age. In addition, it contains a powerful and ubiquitous promoter. To date, more than 1,800 patients have been treated with Zolgensma, with no reports of HCC.
Although there is consensus in the field that additional studies are necessary to better understand the risks associated with AAV integration and that long-term studies are needed to continue to fully evaluate the safety profile of AAV products, the results are controversial. The above shows that the theoretical risk of genotoxicity associated with rAAV chromosomal integration after AAV administration in humans is very low and does not outweigh the potential benefits of this treatment, especially for serious and life-threatening conditions.
References:
- Anguela XM, High KA. Hemophilia B and gene therapy: a new chapter with etranacogene dezaparvovec. Blood Advances. 2024 Apr 9;8(7):1796-803.
- Pipe SW, Leebeek FW, Recht M, Key NS, Castaman G, Miesbach W, Lattimore S, Peerlinck K, Van der Valk P, Coppens M, Kampmann P. Gene therapy with etranacogene dezaparvovec for hemophilia B. New England Journal of Medicine. 2023 Feb 23;388(8):706-18.
- Pipe SW, Leebeek FW, Recht M, Key NS, Castaman G, Miesbach W, Lattimore S, Peerlinck K, Van der Valk P, Coppens M, Kampmann P. Gene therapy with etranacogene dezaparvovec for hemophilia B. New England Journal of Medicine. 2023 Feb 23;388(8):706-18

Zeynab Mirnezhad
Department of Medical Laboratory Sciences. Varastegan Institute for Medical Sciences, Mashhad, Iran
part 3
Immunology
- Mesenchymal stem cells and their application in transplantation therapies
Mesenchymal stem cells and their application in transplantation therapies
INTRODUCTION
Mesenchymal stem cells (MSCs) are vital in regenerative medicine due to their ability to differentiate into various cell types and modulate immune responses. With sources like bone marrow, adipose tissue, and umbilical cord blood, MSCs are easily accessible for clinical use. They promote tissue repair, angiogenesis, and wound healing. Their role in transplantation is particularly significant, as they aid in reducing graft rejection and enhancing transplant success. This review examines how MSCs can transform transplantation therapy through their beneficial mechanisms.
MSCs
Mesenchymal stem cells (MSCs) are exceptionally versatile. Beyond their capacity to differentiate into various cell types, including those from mesodermal, ectodermal, and endodermal origins, MSCs excel as immune regulators. These cells suppress immune responses by inhibiting T cell proliferation, B cell function, and natural killer cell activity while hindering dendritic cell maturation. MSCs achieve this through both direct cell-to-cell interactions and the secretion of anti-inflammatory factors. Although their immunomodulatory effects are potent, it's essential to note that MSC behavior is influenced by the surrounding inflammatory environment. Moreover, MSCs can migrate towards sites of inflammation and tissue damage. This homing ability, influenced by chemokine gradients and receptor interactions, positions MSCs as potential delivery vehicles for therapeutic agents. While their migratory capacity is promising, a deeper understanding of the underlying mechanisms is necessary to fully harness their potential.
Hematopoietic stem cell transplantation (HSCT) remains a challenging therapeutic option due to complications such as graft failure and graft-versus-host disease (GVHD). MSCs have emerged as potential therapeutic agents to address these challenges. By modulating immune responses and potentially promoting engraftment, MSCs offer promise in improving HSCT outcomes. While significant progress has been made, particularly in managing GVHD, further research is needed to fully realize the therapeutic potential of MSCs in this field.
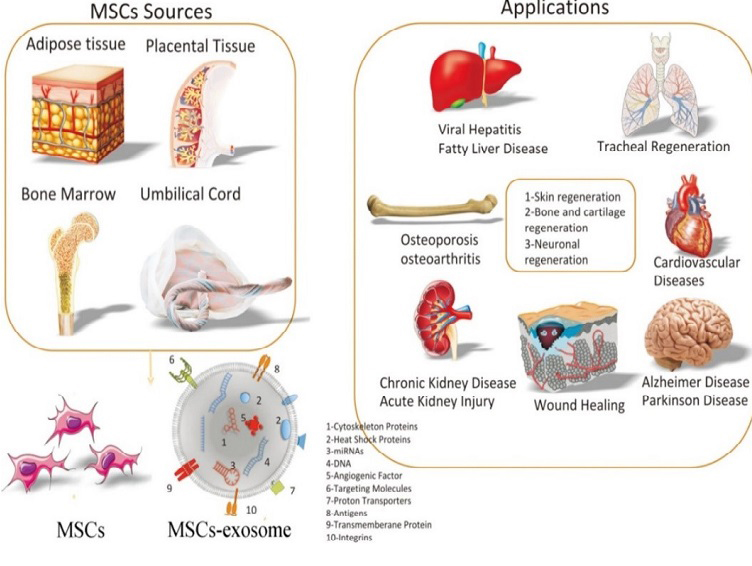
Cardiovascular diseases
Despite advancements in medical care, cardiovascular diseases remain a leading cause of mortality worldwide. Mesenchymal stem cells (MSCs) have emerged as potential therapeutic agents due to their regenerative and immunomodulatory properties. While preclinical studies suggest beneficial effects on cardiac repair, clinical trials have shown mixed results. Further research is necessary to determine the optimal administration route, timing, and dosage of MSCs to achieve sustained cardiac improvement.
Autoimmune diseases
Autoimmune diseases pose significant challenges due to their complex pathophysiology and limited treatment options. MSCs have emerged as potential therapeutic agents due to their immunomodulatory properties. While promising results have been observed in certain autoimmune conditions, such as Crohn's disease, the efficacy of MSC therapy in other diseases, like multiple sclerosis and systemic lupus erythematosus, remains inconsistent. Further research is needed to optimize MSC-based treatments and identify patient populations most likely to benefit.
Liver
Liver diseases, such as cirrhosis, pose significant therapeutic challenges. MSCs offer potential benefits because of their regenerative and immunomodulatory properties. Although initial clinical trials have demonstrated promise in improving liver function, larger-scale studies are needed to confirm the effectiveness and safety of MSC-based therapies. Addressing challenges associated with MSC delivery, engraftment, and retention in the liver will be crucial for maximizing treatment outcomes.
Cancer
Cancer remains a significant global health burden despite advancements in treatment. MSCs have emerged as potential therapeutic agents due to their ability to target tumors and modulate immune responses. Although preclinical studies have shown promise, the translation of MSC-based therapies into clinical practice is still in its early stages. Addressing safety concerns and optimizing MSC engineering for specific cancer types are crucial for realizing the full potential of this approach.
Unorthodox clinical trials showcasing the versatility of MSCs have emerged, exploring the cell's potential in diverse and unexpected applications. While some trials lack a strong scientific foundation, others may reveal unanticipated therapeutic benefits. For instance, MSC-derived stromal vascular fraction has been investigated for vocal fold scarring, and MSC-derived exosomes are being explored for severe lung diseases and dry eye syndrome. These studies highlight the expansive potential of MSCs but also underscore the need for rigorous scientific evaluation and larger-scale trials to establish efficacy and safety. Despite the promising potential of MSCs, significant challenges remain, including inconsistent clinical outcomes, difficulties in standardizing cell manufacturing processes, and concerns about long-term safety. To fully realize the therapeutic potential of MSCs, future research should focus on elucidating underlying mechanisms, developing predictive biomarkers, and optimizing cell delivery methods.
CONCLUSION
Mesenchymal stem cells (MSCs) have great potential for treating various diseases, including cardiovascular and autoimmune conditions, through tissue regeneration and immune response modulation. Despite encouraging initial findings, challenges such as inconsistent outcomes and the necessity for standardized production remain. Future research should prioritize the development of distinct MSC subtypes, refinement of delivery techniques, and comprehension of their mechanisms of action to fully harness their therapeutic capabilities and improve patient outcomes.
References:
- Maqsood M, Kang M, Wu X, Chen J, Teng L, Qiu L. Adult mesenchymal stem cells and their exosomes: Sources, characteristics, and application in regenerative medicine. Life Sci. 2020 Sep 1;256:118002. doi: 10.1016/j.lfs.2020.118002. Epub 2020 Jun 29. PMID: 32585248.
- Umberto Galderisi. Gianfranco Peluso. Giovanni Di Bernardo. Clinical Trials Based on Mesenchymal Stromal Cells are Exponentially Increasing: Where are we in recent years? Stem Cell Reviews and Reports (2022). 29 July 2021/ Published online: 16 August 2021
Ayca Koca Tanriverdi. Onur Polat. Murat Elcin. European Journal of Trauma and Emergency Surgery. 28 February 2019

Arezoo Ezanlo
Department of Medical Laboratory Sciences. Varastegan Institute for Medical Sciences, Mashhad, Iran

Melika Hosseinpour
Department of Medical Laboratory Sciences. Varastegan Institute for Medical Sciences, Mashhad, Iran
part4
Microbiology
A new antibiotic that has nothing to do with good bacteria
Introduction:Bottom of Form
Antibiotics offer a range of advantages; however, they also present considerable limitations stemming from their non-selective mechanism of action. These agents can eradicate both beneficial and pathogenic bacteria within the host organism. The administration of antibiotics can potentially disturb the gastrointestinal tract and its associated microbial ecosystem, which may result in the overproliferation of specific bacterial or fungal species. Gram-negative infections are frequently linked to healthcare-associated infections (HAIs), which are associated with elevated rates of morbidity and mortality. A major obstacle in the treatment of Gram-negative infections is the rising incidence of antibiotic-resistant strains.
Numerous Gram-negative bacteria have developed resistance mechanisms that complicate treatment options. Of particular concern are multidrug-resistant (MDR) strains, including carbapenem-resistant Enterobacteriaceae (CRE) and organisms that produce extended-spectrum beta-lactamases (ESBL). Most antibiotics that have received clinical approval are either effective solely against Gram-positive bacteria (Gram-positive-only antibiotics) or possess broad-spectrum activity, targeting both Gram-positive and Gram-negative bacteria.
Conversely, statistical analyses indicate that the gut microbiome may contain up to 47 percent gram-negative bacteria. Consequently, the use of compounds that non-selectively target and eliminate gram-negative bacteria is likely to result in considerable gut dysbiosis. For instance, colistin, one of the few antibiotics specifically approved for the treatment of gram-negative infections, has been associated with adverse effects such as Clostridium difficile-associated diarrhea and pseudomembranous colitis, which can be life-threatening. Additionally, colistin poses toxic risks to hepatic and renal functions, leading to its designation as an antibiotic of last resort. There is an urgent need for the development of antibiotics that selectively target gram-negative bacteria while sparing gram-positive bacteria, as well as for distinguishing pathogenic bacteria from commensal organisms.
Lolamicin and clinical trials:
In addressing the numerous challenges related to the indiscriminate targeting of gram-negative bacteria, a research team from the University of Illinois at Urbana-Champaign investigated a collection of pharmaceuticals developed by AstraZeneca. These agents function by obstructing the Lol system, which is a lipoprotein transport mechanism exclusive to gram-negative bacteria and exhibits genetic diversity among both pathogenic and beneficial microorganisms. The Lol system comprises five unique proteins, designated LolA through LolE, with three of these proteins forming the LolCDE transporter complex. The efficacy of these drugs against gram-negative infections was not observed unless the researchers first compromised critical bacterial defense mechanisms in a controlled laboratory setting.
In a series of experimental investigations, researchers developed structural modifications of the Lol inhibitors and assessed their efficacy against gram-negative and gram-positive bacteria in cell culture environments. Among the newly synthesized compounds, Lolamicin demonstrated selective activity against certain laboratory strains of gram-negative pathogens, including E. coli, K. pneumoniae, and E. cloacae. The findings indicated that Lolamicin exhibited no significant impact on gram-positive bacteria within the cell culture setting. Notably, at elevated concentrations, Lolamicin was capable of eliminating up to 90 percent of multidrug-resistant clinical isolates of E. coli, K. pneumoniae, and E. cloacae.
Research has demonstrated that antibiotics inducing gut dysbiosis in murine models exhibit similar effects in humans. In a study involving the oral administration of Lolamicin to mice suffering from drug-resistant septicemia or pneumonia, the results indicated a complete survival rate of 100 percent in the septicemia cohort and a survival rate of 70 percent in the pneumonia cohort, as reported by the research team. Furthermore, comprehensive investigations were conducted to assess the impact of Lolamicin on the gut microbiome. The murine microbiome serves as an effective model for studying human infections due to the significant similarities between the gut microbiomes of humans and mice.
The research findings indicated that the administration of the antibiotics amoxicillin and clindamycin resulted in significant alterations in the overall composition of bacterial populations within the murine gastrointestinal tract, leading to a reduction in the prevalence of several beneficial microbial taxa. Conversely, treatment with Lolamicin did not induce any substantial modifications in taxonomic diversity during the three-day treatment period or in the subsequent 28-day recovery phase. In a follow-up experiment involving mice, the researchers demonstrated that subjects exposed to Clostridium difficile following Lolamicin treatment were capable of effectively eliminating the pathogen, exhibiting minimal to no colonization by C. difficile. In contrast, mice that received amoxicillin or clindamycin were unable to eradicate C. difficile and exhibited substantial colonization by the pathogen. The preservation of the gut bacterial community in mice treated with Lolamicin appears to facilitate the successful clearance of C. difficile colonization, thereby distinguishing Lolamicin from currently utilized antibiotics in clinical practice.
Conclusion:
According to Hergenrother, the development of antibiotics that effectively target pathogenic microorganisms while preserving beneficial gut bacteria is feasible for treating gram-negative infections, which are among the most difficult to manage. The authors of the study concluded that "Lolamicin demonstrates activity against a diverse array of over 130 multidrug-resistant clinical isolates, exhibits efficacy in various murine models of acute pneumonia and septicemia, and maintains the integrity of the gut microbiome in mice, thereby preventing secondary infections with C. difficile." The intestinal microbiome plays a crucial role in sustaining host health, and its disruption can lead to numerous adverse outcomes, including infections caused by C. difficile and other complications. Therefore, the development of pathogen-specific antibiotics, such as Lolamicin, is essential for reducing collateral damage to the gut microbiome; this microbiome-sparing characteristic would render such antibiotics more advantageous for patients compared to those currently utilized in clinical settings.
References:
- McDonald LC, Young VB, Wilcox MH, Halpin AL, Chaves RL. Public health case for microbiome-sparing antibiotics and new opportunities for drug development. mSphere. 2024 Aug 28;9(8):e0041724. doi: 10.1128/msphere.00417-24. Epub 2024 Aug 2. PMID: 39092918; PMCID: PMC11351033.
- McDonald LC, Young VB, Wilcox MH, Halpin AL, Chaves RL. Public health case for microbiome-sparing antibiotics and new opportunities for drug development. mSphere. 2024 Aug 28;9(8):e0041724. doi: 10.1128/msphere.00417-24. Epub 2024 Aug 2. PMID: 39092918; PMCID: PMC11351033.

Mahdieh Sadat Hosseini Nezhad
Department of Medical Laboratory Sciences. Varastegan Institute for Medical Sciences, Mashhad, Iran
part 5
Genetics & Biotechnology
- Investigation of the Role of Effective Gene Therapy in Lung Cancer Control and Recovery
-Potential use of cancer cells as expression systems in pharmaceutical biotechnology
-Spatial Transcriptomics in Neuroscience and Neurological Disorders
Potential use of cancer cells as expression systems in pharmaceutical biotechnology
Introduction to Cancer Cells as Expression Systems
Many pharmaceuticals are being used worldwide to treat various medical conditions every day. Pharmaceutical biotechnology companies play a significant role in drug manufacturing. Scientists have developed numerous techniques to create these chemicals. One such technique involves inserting the gene or cDNA of a desired protein into specific expression systems, from which the resulting products can be extracted using various methods. For example, producing liposomes is one of these techniques. One of the trending topics is the utilization of pharmaceutical biotechnology in treating diseases, especially cancer. Cancer cells are carriers that encapsulate chemotherapeutic drugs and are crucial in anticancer therapy, as they are derived from the host cell membrane. This approach has the potential to significantly reduce systemic toxicity, enhance targeting, and improve drug penetration into tumor cells, thereby enhancing its clinical application in the future. Research and advancements have focused on encapsulating pharmaceuticals using erythrocytes, white blood cells, platelets, immune cells, and even tumor cells.
Cultivation and testing of cancer cells
Biologic substances are produced by scientists by introducing a specific protein's gene or cDNA into well-characterized expression systems, then extracting the result and processing it further to create medicine or diagnostic tools. For instance, due to their ability to properly fold, assemble, and modify proteins after translation, cultivated cell lines from mammals have become the predominant technology for producing recombinant proteins for medical use. Therefore, when a protein is expressed in mammalian cells compared to other hosts like bacteria, plants, or yeast, it can have higher quality and effectiveness. However, cancer cells are currently being explored as expression systems in pharmaceutical biotechnology due to their unique characteristics, such as their capacity for intricate post-translational modifications. Another distinguishing feature is their ease of development. Because cancer cells divide more frequently than normal cells, they are easier to culture. Additionally, normal cells have shorter telomeres that shorten with each cell division, while cancer cells survive longer in the culture process because their telomeres persist. Moreover, certain cancer cells can produce biological chemicals that can be used as medications, such as growth hormones, cytokines, antibodies, and more.
Case Studies: Cancer Cell Lines in Biopharmaceutical Production
Cancer cell lines are now widely utilized as tools, especially in the field of producing therapeutic antibodies and proteins. The NS0 myeloma cell line, known for its ability to synthesize proteins in large quantities, is commonly employed in the production of monoclonal antibodies. Researchers have successfully utilized the NS0 cell line to produce a chimeric B72.3 IgG4 antibody, achieving a production rate of 10–۱۵ pg/cell/day and reaching 560 mg/L in a fed-batch fermentation system. The productivity and stability of expression without pressure exhibited by cancer cell lines demonstrate their potential for large-scale production of biopharmaceuticals. Another notable example is the HeLa cell line, which has been a crucial tool in cancer research for over 50 years. Due to its robust growth characteristics and ability to undergo extensive post-translational modifications, the HeLa cell line is valuable for producing materials like vaccines and diagnostic tools, showcasing its versatility. These examples highlight how cancer cell lines play a significant role in advancing biotechnology by providing reliable platforms for the production of medicinal products.
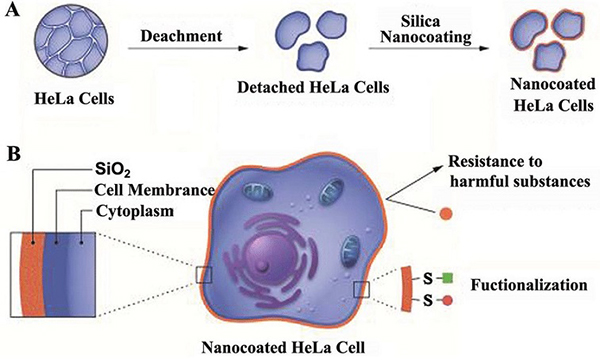
Ethical and Regulatory Challenges
Typically, in any course of treatment, there are adverse effects and other challenges that can complicate the whole procedure. Here are five challenges of using cancer cells in pharmaceutical biotechnology.
- Genetic and biological diversity: The majority of cancer cells exhibit genetic and biological variations, which complicates the process of conducting experiments. Consequently, additional standards and careful cell type selection are required.
- Cell Stability: Some cancer cell lines are typically unstable over time and may show significant variability. These modifications can impact the quality and reproducibility of the results.
- Ethical issues: On the other hand, using cancer cells would require higher ethical and legal standards if they were obtained from actual patients as opposed to cell lines that can be continuously cultivated in laboratories.
- Low efficiency: It is possible that cancer cells do not always effectively express the required proteins, leading to increased medication costs and slower production.
- Unrealistic modeling: Not every cancerous state or characteristic can be replicated in vivo, making it challenging to generalize experimental results.
Future Prospects and Innovations
These are the four most significant emerging opportunities for utilizing cancer cells in the production of biopharmaceuticals:
- Technological improvements in the field: Although CRISPR technology cannot reduce the genetic instability of cancer cells and enable more precise control of gene expression, it can pave the way for the introduction of more effective methods for modifying the DNA sequence.
- Innovations in treatment: Modifying the genes of a cancer patient could lead to personalized therapy that is more efficient and accurate compared to the current standard treatment methods.
Bioinformatics and data mining usage: The combination of data mining with machine learning can assist in identifying patterns and predicting outcomes related to exploiting cancer cells.
In the future, we will have the capability to develop mobile cellular factories. These factories will provide us with new tools to generate cells and potentially cure cancer. They will possess the ability to detect and eliminate diseased cells.
Conclusion
Even though the use of cancer cells in pharmaceutical biotechnology presents challenges that need to be addressed, most of these issues can be resolved through advancements in science and technology. A future where cancer cells are utilized to standardize expression systems for drug production will necessitate ongoing monitoring to ensure safety, ethical standards, and legal compliance. However, with the progress of new technologies and ongoing research, these possibilities appear more promising than ever.
References
- Du Y, Wang S, Zhang M, Chen B, Shen Y. Cells-based drug delivery for cancer applications. Nanoscale research letters. 2021 Dec;16:1-0.
- O’Flaherty R, Bergin A, Flampouri E, Mota LM, Obaidi I, Quigley A, Xie Y, Butler M. Mammalian cell culture for production of recombinant proteins: A review of the critical steps in their biomanufacturing. Biotechnology advances. 2020 Nov 1;43:107552
- Vredevoogd DW, Peeper DS. Heterogeneity in functional genetic screens: friend or foe?. Frontiers in Immunology. 2023 Jun 16;14:1162706.
- Yao S, Jin B, Liu Z, Shao C, Zhao R, Wang X, Tang R. Biomineralization: From Material Tactics to Biological Strategy. Adv Mater. 2017 Apr;29(14). doi: 10.1002/adma.201605903. Epub 2017 Feb 23. PMID: 28229486.

Nastaran Tavabaki
Department of Medical Laboratory Sciences. Varastegan Institute for Medical Sciences, Mashhad, Iran

Setareh Eskandari
Department of Medical Laboratory Sciences. Varastegan Institute for Medical Sciences, Mashhad, Iran
Examining the impact of efficient gene therapy on the management and rehabilitation of lung cancer
Introduction
Lung Cancer (LC) is the most common cause of death worldwide. Patients with LC are typically diagnosed at advanced stages, and the five-year survival rate for these patients is approximately 16%. Despite decades of research into LC treatments, clinical outcomes remain suboptimal, highlighting the need for the development of new technologies for disease management. Given the role of genetic and epigenetic changes in oncogenes and tumor suppressor genes in cancer progression, gene therapy has emerged as a focal point in cancer treatment research. Compared to conventional methods such as chemotherapy, gene therapy generally has fewer side effects. Unlike traditional gene therapy methods that produce only temporary effects, genetic modification tools can offer more permanent solutions. In recent years, numerous studies have effectively employed the CRISPR-Cas9 approach to modify gene expression in cells. This system induces mutagenesis, facilitates site-specific epigenetic changes, and regulates gene expression. Gene therapy utilizing the CRISPR-Cas۹ technique is rapidly gaining popularity within the scientific community, primarily due to its versatility, cost-effectiveness, and high efficiency. However, while there is a wealth of laboratory-based experiments and findings using CRISPR as a gene-editing tool, the critical question remains: how many of these findings can be translated into actionable strategies to combat specific disease conditions?
Genetics and lung cancer
Many genetic mutations increase the risk of developing cancer. While single and isolated point mutations generally have minor effects, the combination of several mutations can significantly elevate the risk of diseases such as cancer. In LC, multiple mutations in oncogenes and tumor suppressor genes have been observed. Notable genetic changes in LC include the deletion of the short arm of chromosome 3 and the duplication of chromosomes 1 and 12. Loss of chromosome 3 alleles has been observed in approximately 90% of small cell lung cancer (SCLC) cases and 50% of non-small cell lung cancer (NSCLC) cases. The main oncogenes activated in NSCLC are epidermal growth factor receptor, anaplastic lymphoma kinase (ALK), Myc, Bcl-2, and KRAS. Commonly mutated tumor suppressor genes include TP53, p16, and retinoblastoma (RB1). These mutations are associated with poor prognosis in patients. As members of the receptor tyrosine kinase superfamily, EGFR, and ALK contribute to various cellular processes such as proliferation and differentiation. Mutations in these receptors can lead to ligand-independent activity, subsequently causing malignancy. Among the genes mutated in SCLC, the tumor suppressors TP53 and RB1 are nearly universal. Furthermore, methylation or inactivation of the tumor suppressor RASSF1A at 3p21 has been reported in 70–۱۰۰% of SCLCs and 45–۶۵% of NSCLCs. Other genes that may be deleted in the 3p region include RARB, caveolin-1, FHIT, and β-catenin. Additional genetic changes may occur in histone acetyltransferases, including CREBBP and EP300, as well as in the tumor suppressor PTEN, NOTCH family genes, and MYC-related genes.
CRISPR/CAS system
Long periods of research on genome manipulation methods have enabled precise modification of target DNA sequences at the cellular level. CRISPR-Cas9 gene editing technology is a new and high-efficiency method for the effective treatment of various genetic disorders, especially cancers. The CRISPR-Cas system is part of the prokaryotic adaptive defense system against the invasion of plasmids and viral particles. Cas is an endonuclease associated with an RNA derived from CRISPR sequences. This system is activated after exposure to exogenous DNAs of bacteriophages or invading plasmids and can identify and remove invading molecules. In bacteria and archaea, the Cas protein recognizes foreign invading plasmids or phage DNA with the help of sgRNAs that come from CRISPR sequences.
CRISPR/CAS system in cancer
As mentioned above, the invention of CRISPR-Cas9 systems revolutionized genome manipulation processes and increased their efficiency. The application of this system in cancer gene therapy is due to its ability to induce somatic mutations in cells. Indeed, CRISPR-mediated genetic manipulation has led to the discovery of new therapeutic targets in cancer. The combined delivery of Cas9 and multiple sgRNAs can simultaneously induce multiple genetic changes in tumor cells. In this regard, the CRISPR method has contributed to the discovery of MELK, a target molecule that subsequently led to the design of OTS167, a MELK blocker, which is currently being studied in multiple clinical trials. In one study, CRISPR-based disruption of Mll3 in peripheral conditions in mouse-derived hematopoietic stem cells helped uncover the tumor suppressor role of Mll3 in acute myeloid leukemia. In another study, plasmids encoding Cas9 and sgRNA were used to deliver the tumor suppressors Pten and Trp53 into hepatocytes via hydrodynamic gene transfer in vivo. Soda et al used a CRISPR-Cas9 complex to induce oncogenic chromosomal rearrangements in mice. To this end, they delivered an adenovirus containing Cas9 and two sgRNAs to induce Eml4-Alk (echinoderm microtubule-associated protein 4-ALK) inversion. The results confirmed that CRISPR-based manipulation of the body's genome can effectively induce cancer. The CRISPR system can also be used to detect mutations responsible for treatment resistance in cancer. For example, NAMPT, which encodes nicotinamide phosphoribosyltransferase and is the target of the anticancer agent KPT-۹۲۷۴, was identified thanks to CRISPR technology. CRISPR-based mutagenesis studies have also led to the identification of MEK and BRAF variants that confer resistance to the anticancer drugs selometinib and vemurafenib, respectively. CRISPR-based rearrangements of CTCF binding sites, frequently mutated in various types of cancers, were shown to alter the activities of their promoters and enhancers as well as gene topology. CRISPR-Cas9 method has been used to inactivate PD.
CRISPR-CAS9 system in LC modeling
LC modeling and tumor cell engineering is one of the most important applications of CRISPR-based techniques. CRISPR-mediated removal of tumor suppressor genes, particularly through the NHEJ process, has provided an opportunity to engineer normal tumor cells. Guernet et al used a CRISPR barcoding strategy to predict different mechanisms of resistance to EGFR inhibitors in NSCLC. First, the CRISPR-Cas9 system has been exploited to insert multiple point mutations into a single genomic region to tag and track tumor cells. To develop a new model of EGFR inhibitor-resistant NSCLC, they generated a specific sgRNA and a single-stranded DNA oligonucleotide (ssODN) containing the T790M mutation. A few additional silent mutations were also inserted to provide a genetic barcode. To avoid misplacing the two DNA sequences, EGFR inhibitor-sensitive PC9 cells were transfected with a CRISPR/Cas9 plasmid encoding Cas9 and a sgRNA, along with one of two ssODNs. Gefitinib treatment was found to increase the ratio of EGFR-T790M to EGFR-T790T, indicating increased resistance to the EGFR inhibitor.
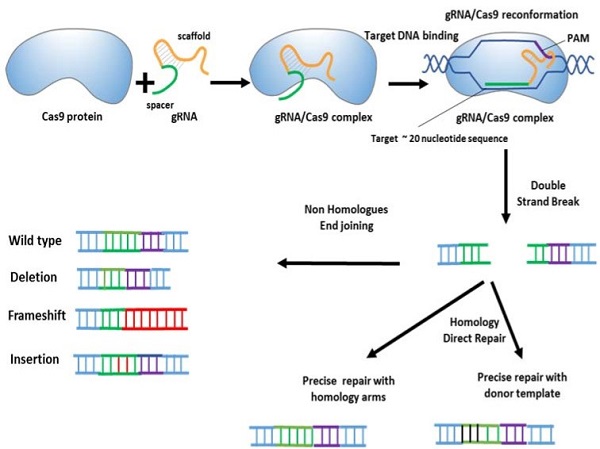
Reference
- Akbari Kordkheyli V, Rashidi M, Shokri Y, Fallahpour S, Variji A, Nabipour Ghara E, Hosseini SM. CRISPR/CAS System, a Novel Tool of Targeted Therapy of Drug-Resistant Lung Cancer. Adv Pharm Bull. 2022 Mar;12(2):262-273. doi: 10.34172/apb.2022.027. Epub 2021 Apr 3. PMID: 35620343; PMCID: PMC9106967.
- Kazemizadeh H, Kashefizadeh A. CRISPR-Cas9-mediated gene therapy in lung cancer. Clin Transl Oncol. 2023 May;25(5):1156-1166. doi: 10.1007/s12094-022-03039-8. Epub 2022 Dec 10. PMID: 36495467.
- Nair, J., Nair, A., Veerappan, S. et al. Translatable gene therapy for lung cancer using Crispr CAS9—an exploratory review. Cancer Gene Ther ۲۷, ۱۱۶–۱۲۴ (۲۰۲۰). https://doi.org/10.1038/s41417-019-0116-8

Ali Ahmadi
M.Sc. Student of Genetics, Department of Genetics, Faculty of Advanced Technologies and Science, Islamic Azad University Tehran Medical Science, Tehran, Iran
Spatial Transcriptomics in Neuroscience and Neurological Disorders
Introduction:
Spatial transcriptomics is an innovative technique that helps researchers understand tissues and cellular processes by providing information about gene activity in three-dimensional space. This approach allows scientists to visualize where specific genes are expressed within tissues, offering insights into how cells function and interact in their natural environment.
What is spatial transcriptomics?
Spatial transcriptomics is a research technique that combines gene expression analysis with the spatial arrangement of cells within a tissue. It allows scientists to map the locations of specific RNA molecules in a tissue sample, providing insights into how genes are expressed in different regions. This technique helps researchers understand the complex interactions between cells, the architecture of tissues, and how these factors relate to health and disease.
Spatial Transcriptomics in Neuroscience
Spatial transcriptomics is a technique used in neuroscience to examine gene expression in specific locations within brain tissue. Unlike traditional transcriptomics, which analyzes RNA from an entire tissue sample, spatial transcriptomics combines gene expression data with the spatial information of where those genes are expressed within the tissue.
This approach allows researchers to understand how different brain regions function, how cells interact, and how molecular changes correlate with various neurological conditions or brain development. By mapping gene activity to specific locations, scientists gain insights into the complex cellular landscapes of the brain, which helps advance our understanding of neural processes and disorders.
Impact on Research on Neurological Disorders
Degenerative disorders, which involve the progressive decline of tissue or organ function (such as Alzheimer's or Parkinson's), can potentially see significant improvements through spatial transcriptomics. This technology enables scientists to study gene activity within specific locations in tissue samples, providing insights into how these disorders develop and progress. By comprehending the spatial organization of gene expression, researchers can better identify disease mechanisms and potentially develop targeted treatments.
Bridging the Gap Between Research and Clinical Practice
Accurately identifying different types of cells is crucial for developing treatments for specific health conditions. Nociceptors, which are responsible for detecting pain, are particularly important targets for managing both short-term (acute) and long-term (chronic) pain. Understanding the gene expression patterns (transcription profiles) of these cells can help researchers select appropriate animal models for experiments and may provide insights into identifying potential biomarkers for pain treatment.
Spatial transcriptomics can be utilized in personalized medicine by analyzing gene activity in specific locations within tissues. This approach enhances our understanding of how individual patients' diseases develop and respond to treatments. By tailoring therapies based on the unique cellular environment of each patient's tissues, doctors can offer more effective and targeted treatments.
Conclusion: The Future of Spatial Transcriptomics in Medicine
Spatial transcriptomics is an innovative technique that combines two fields: spatial biology and genomics. It allows scientists to map gene activity in tissues while preserving their spatial context. Here is a concise overview of its future applications in medicine:
- Personalized Medicine
- Cancer Research
- Neuroscience
- Drug Development
- Regenerative Medicine
Overall, spatial transcriptomics holds great promise for advancing medical research and improving patient care by providing a detailed understanding of biological processes in their natural context.
Reference
- Jung N, Kim TK. Spatial transcriptomics in neuroscience. Exp Mol Med. 2023 Oct;55(10):2105-2115. doi: 10.1038/s12276-023-01093-y. Epub 2023 Oct 2. PMID: 37779145; PMCID: PMC10618223.
- Zhang L, Xiong Z, Xiao M. A Review of the Application of Spatial Transcriptomics in Neuroscience. Interdiscip Sci. 2024 Jun;16(2):243-260. doi: 10.1007/s12539-024-00603-4. Epub 2024 Feb 20. PMID: 38374297.

Maryam Rohbakhsh Faalnezhad
Department of Medical Laboratory Sciences. Varastegan Institute for Medical Sciences, Mashhad, Iran
part 6
News
Parasites found in cat feces may have the potential to treat serious conditions such as Alzheimer's and Parkinson's disease
The difficulty of administering precise therapeutic interventions across the blood-brain barrier and into particular neuronal populations has been a persistent issue in neurological healthcare. Toxoplasma gondii, a parasite commonly linked to feline excrement, has exhibited a notable capacity to traverse this barrier. This organism naturally migrates from the gastrointestinal tract to the central nervous system, secreting proteins into neuronal cells.
The research team concentrated on the delivery of the MeCP2 protein, which is recognized for its therapeutic potential in treating Rett Syndrome, a serious neurological disorder resulting from mutations in the MECP2 gene. The genetically modified parasites effectively synthesized and transported the MeCP2 protein to the appropriate cellular sites within brain organoids and murine models.
The researchers intend to conduct additional experiments to confirm that the engineered parasites are programmed to perish following the delivery of their therapeutic payload, thereby reducing the risk of potential adverse effects. This measure is essential for preventing further cellular damage and ensuring the safety of the treatment.
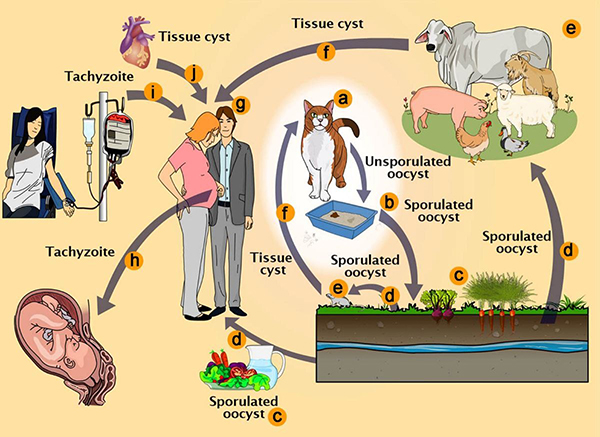
Reference
- Attias, M., Teixeira, D.E., Benchimol, M. et al.The life-cycle of Toxoplasma gondii reviewed using animations. Parasites Vectors ۱۳, ۵۸۸ (۲۰۲۰). https://doi.org/10.1186/s13071-020-04445-z
- interestingengineering.com

Mahdieh Sadat Hosseini Nezhad
Department of Medical Laboratory Sciences. Varastegan Institute for Medical Sciences, Mashhad, Iran
Can a blood test predict diabetes, kidney disease, Alzheimer's, and other age-related diseases?
A recent study in Nature Medicine indicates that the proteome might accurately indicate a person’s biological age. The proteome refers to the entire collection of proteins produced by an organism. The research aimed to determine whether scientists could create a “proteomic aging clock” that would help to predict the risk of some common age-related diseases.
By using proteomics data, it is possible to assess aging by comparing an individual’s biological function to what is expected for their chronological age. Although most biological aging clocks depend on DNA methylation, examining protein levels may provide a more straightforward insight into the mechanisms of aging, particularly since proteins are central to drug development.
“This could shift the focus from treating diseases after they occur to predicting and preventing them before they start,” said Norman, a board-certified geriatric nurse practitioner with the National Council on Aging, adding, “This information is another tool in the tool belt that healthcare providers can use to help people better understand their health and choices.”
Reference

Nazanin Zeinab Arefipour
Department of Medical Laboratory Sciences. Varastegan Institute for Medical Sciences, Mashhad, Iran

